ESPCI- Light Sheet Fluorescence Microscopy project
[Interview with]
Emna, Xiangdong, and Lilian, engineering students at ESPCI
ight Sheet Fluorescence Microscopy project with the LBX-648 from Oxxius
[Interview with]
Emna, Xiangdong, et Lilian, engineering students at ESPCI

Team members (Emna, Xiangdong, Lilian)
Hi, could you introduce yourselves and give us a quick understanding of what ESPCI is?
Hi ! We are Emna, Xiangdong and Lilian, 3 Master students at ESPCI Paris-PSL. ESPCI is a prestigious engineering school specializing in physics and chemistry, as well as biology. The training provided there is very multidisciplinary and research-oriented. Half of the training time is devoted to practical work, carried out in the laboratories of the school, which is also a research institute.
What kind of research do you carry out ?
In addition to the practical work, we conduct a Scientific Team Project. As part of this project, our trinominal is working on the development of a light sheet microscope (called LSFM for Light Sheet Fluorescence Microscopy, or SPIM, for Selective Plane Illumination Microscopy) for application in biology. The objective is to produce an optical setup of fluorescence microscopy to illuminate a biological sample with a thin sheet of light. This method makes it possible to acquire an image of a plane of the sample much faster than confocal microscopy, where the image of a plane is obtained point by point. It is therefore suitable for the observation of phenomena requiring good temporal resolution (observation of the response to stimulation in neuroscience, for example). It also has the advantage of less exposure of the sample to laser light, thus inducing less phototoxicity and photobleaching of the fluorophores. We then applied our setup to the observation of Drosophila pupae, an organism widely used for neuroscience research. It is therefore an interdisciplinary project, at the interface between technological development and its application in primary research in biology.
What was your research process and what were the critical points?
To build our setup, we were inspired by the work published on light sheet microscopy, and in particular from the online platform OpenSPIM, which gives advice and an example of light sheet microscopy setup for non-specialists.
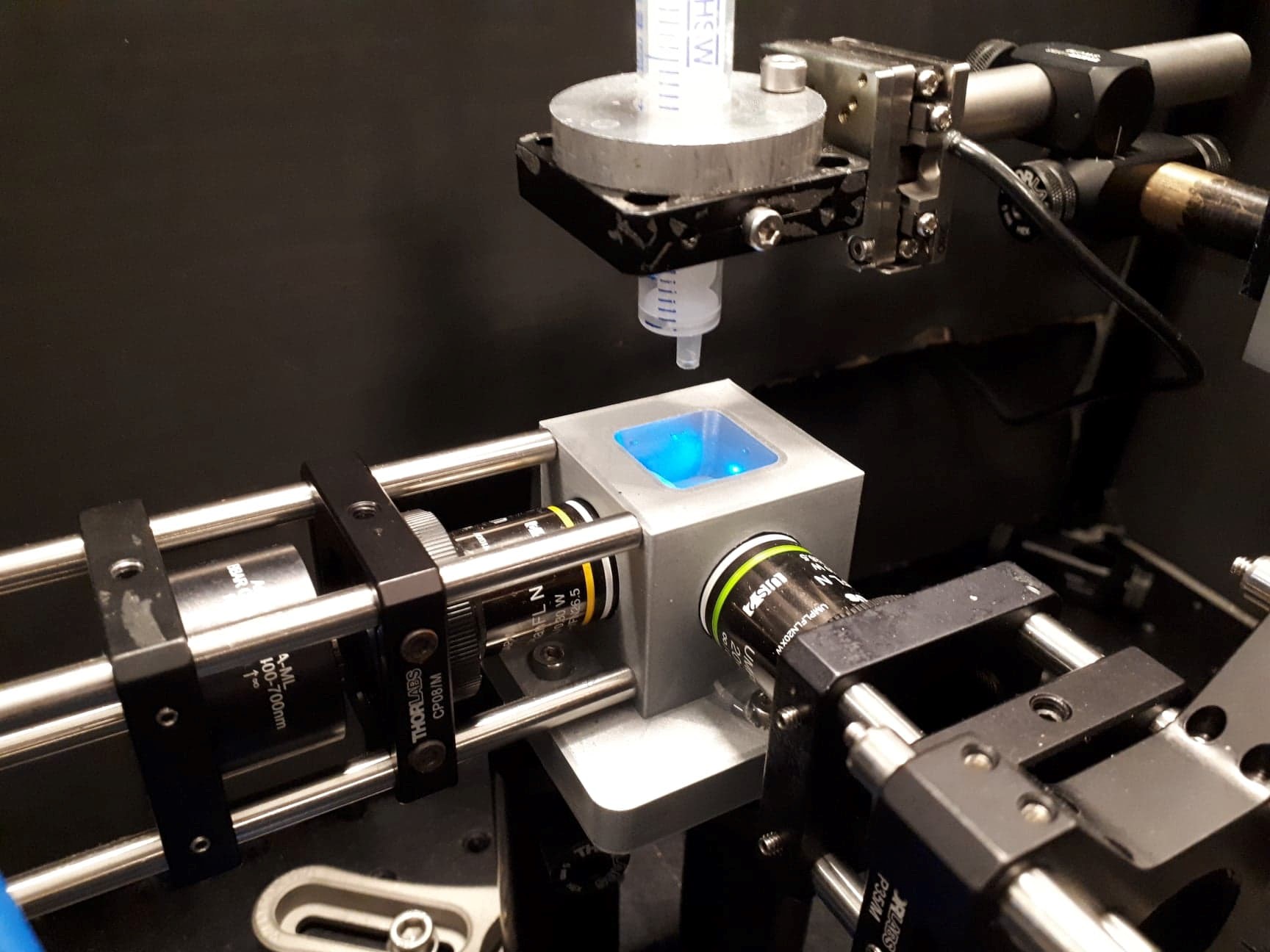
The light sheet is obtained by the inclusion of a cylindrical lens in the optical setup. The fluorescence emission is collected perpendicular to the laser sheet. Once the optical setup was done, the most delicate step was to develop a mounting system for the biological sample to be observed. The Drosophila pupa, embedded in an agarose gel, is suspended from the end of a syringe submerged in water (the objectives used for illumination and detection are water dipping objectives).
We used translation stages so that we could move our sample in the 3 directions of space.
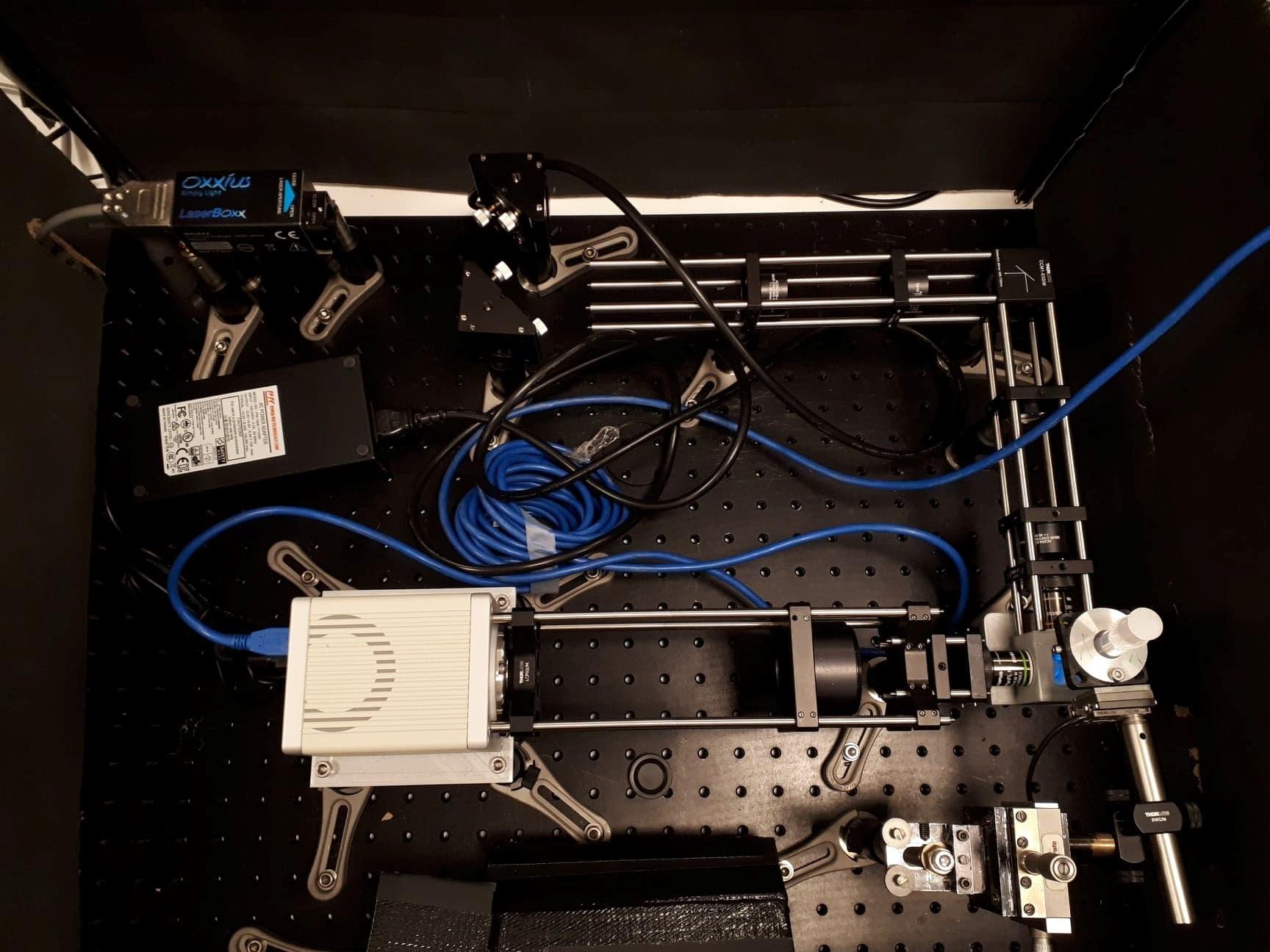
credit: ©Optical setup seen from above ESPCI, 2021

credit: ©Mounting system for the biological sample, between the 2 objectives (perpendicular to each other) ESPCI, 2021

credit: ©Drosophila pupa mounted in agarose, suspended from the end of a syringe ESPCI, 2021
Why did you choose the LBX-488 from Oxxius ?
The goal of our project was to observe Drosophila pupae expressing GFP (Green Fluorescent Protein), a fluorescent marker widely used in biology. Therefore, we needed a laser light source with a power suitable for the illumination of biological samples, at the optimum excitation wavelength of GFP. The LBX 488 from Oxxius was perfectly suited to our needs, which is why we selected it for our project.
ZOOM ON |
LBX – 488 |
 Oxxius LBX 488 Laser source used in our setup |
Emission wavelength : 488 nm (+/- 5 nm) Linewidth : ≤2 nm Output power : from 40 to 200 mW Power stability: + 0.5% Power adjustment range : 0-100% Optical noise : ≤ 0.2% Beam waist diameter : 0.7mm Beam divergence : ≤1.2 mrad Beam quality factor: ≤1.25M2 Beam circularity : ≥90% Beam pointing stability : ≤ 5 µrad/K Polarization state / linear, vertical at +/-5° Digital modulation: Max. modulation frequency: 150Mhz Analog modulation : Bandwidth ≥3 MHz |
Do you have any results to share with us ?
At the end of our project, we were able to obtain images of Drosophila pupae with our light sheet microscopy setup.
The acquisition of images on several planes also allowed us to produce 3D images slice by slice.
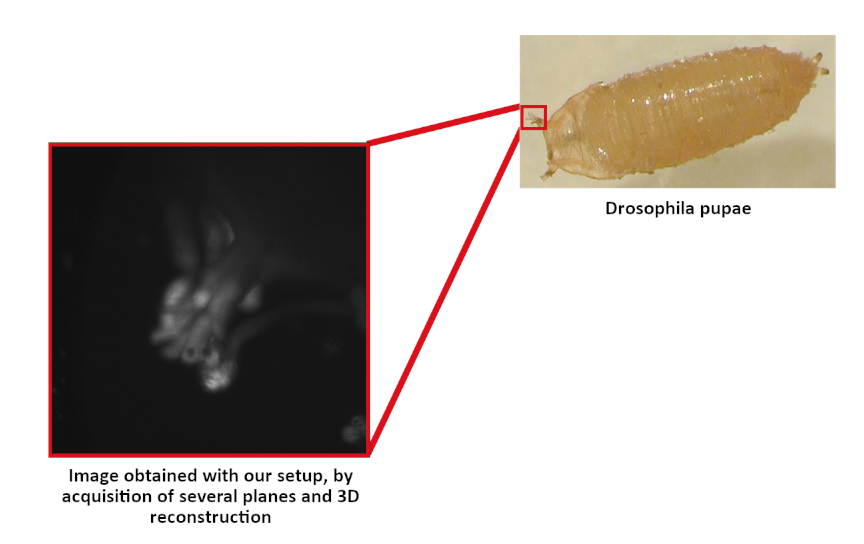
The stacked 2d images were evenly spaced at different depths of the sample.
We then used the ImageJ software to define the distance between each image plane, and reconstruct a 3D image from the stack of 2D images.
Our project may be used and improved by future graduate students to observe biological samples.
Application areas include developmental biology (e.g. observing the development of a fluorescently labelled organ in a larva), or neuroscience (e.g. observing the response of a Drosophila brain area to a stimulus).

